简单总结
这是 OpenAI 的 12 Days 第二天直播,主要介绍了强化微调 (Reinforcement Fine-Tuning) 技术。这项技术允许用户使用强化学习来定制模型, 使其在特定领域内达到专家水平,如果你要想试试的话,可以填一下 OpenAI 的 RFT 申请表单。原视频链接:https://www.youtube.com/watch?v=yCIYS9fx56U
亮点
-
🚀 强化微调是一种新的模型定制方式,不同于传统的监督微调,它不仅是模仿输入, 而是教会模型以新方式思考。适用于法律、金融、医疗等需要专业知识的领域。提供了完整的训练基础设施和评估工具,计划在 2024 年初向公众推出,特别适合拥有专家团队的研究机构,可以显著提升模型在特定任务上的表现,可以帮助科研人员更好地理解和处理专业领域问题
-
🧬 视频也展示了在罕见遗传病诊断领域的应用案例,只需几十个示例就可以让模型学会在特定领域进行有效推理,使用评分器对模型输出进行评估, 分数范围 0-1,微调后的 O1 mini 模型性能显著提升, 超过了基础 O1 模型
如果你从事研究、企业级别或需要自定义 AI,比如筋斗云 SEO 有较多自研技术,这可能有用。 但是,对于大多数用户来说,这是高级技术的一部分并且代价高昂,因此大多数用户可能不会或不需要使用 RFT。
强化微调技术中文版图文
大家好,我是 Mark,OpenAI 的研究主管。昨天,我们正式发布了 O1 模型,它已经可以在 ChatGPT 中使用,API 版本也即将上线。

如果您还不了解 O1,它是我们最新的模型改进系列,允许模型在回答问题前先进行思考。今天,我们将预览一项更强大的模型定制化技术:强化微调。这项技术能让用户使用自己的数据集来微调 O1 模型。
需要强调的是,这可不是普通的微调。强化微调 利用了强化学习算法,就像当初我们利用它将模型的水平从高中生提升到博士专家一样。这项技术将于明年正式发布,但如果您是高校、研究人员或企业用户,我们稍后会提供参与我们测试项目的信息。
为什么要使用强化微调呢?因为它可以让您 将自己的 “黄金数据集” 转化为独特的模型,就像我们一样,但服务于您自己的用户和客户。接下来,John、Julie 和 Justin 会详细介绍这项技术。
John:大家好,我是 John Allard,OpenAI 的工程师。
Julie:大家好,我是 Julie W,OpenAI 的研究员。
Justin:我是 Justin Ree,伯克利实验室的计算生物学家。
强化微调:打造 AI 专家
John:今天,我们很高兴向大家介绍一种全新的 O1 模型定制化方法:强化微调(RFT)。开发者、研究人员和机器学习工程师 将首次能够使用强化学习来创建 “专家模型”,使其能够在特定领域内出色地完成任务。
我们相信任何需要 AI 模型具备深厚专业知识的领域都能从中受益。如果您从事 法律、金融、工程或保险 行业,这项技术将是您的得力助手。例如,我们最近与 Thomson Reuters 合作,使用 RFT 微调了 O1 Mini,使其成为他们联合咨询 AI 中的法律助理。这个工具可以帮助法律专业人士完成最具分析性的工作流程。
强化微调 vs. 监督微调
Julie:部分朋友可能比较熟悉我们去年初发布的 监督微调 API。监督微调的优势在于它可以让模型 复制输入文本或图像中的特征。如果您想改变模型的语气、风格或回复格式,监督微调非常有效。
而我们今天介绍的 强化微调,或者我应该称之为 “驯鹿强化” 微调,则完全不同。您不仅仅是教模型模仿输入,而是教它以全新的方式对自定义领域进行推理。
当模型遇到问题时,我们会给它时间进行思考,然后 对它的最终答案进行评分。利用强化学习,我们会 强化那些得出正确答案的思路,并抑制那些得出错误答案的思路。只需几十个例子,模型就能学会以新的有效方式进行推理。
John:难以置信,仅仅 12 个例子就能做到!这可不是普通的微调能做到的。
Julie:没错!对于大型语言模型来说,几十个例子真的微不足道。
强化学习:OpenAI 前沿模型的训练秘诀
John:我们的模型定制化平台将首次支持 强化学习。值得注意的是,我们内部也使用同样的技术来训练 GPT-4 和 O1 系列等前沿模型。这项技术的一个令人兴奋的应用领域是科学研究。为了更好地说明这一点,我们邀请到了 Justin Ree。Justin 是伯克利实验室的研究员,他的研究方向之一是使用计算方法来理解罕见疾病的遗传原因。
Justin,非常感谢您来到这里。您能否详细介绍一下您的研究以及强化微调将如何提供帮助?
强化微调助力罕见疾病研究
Justin:谢谢,很高兴来到这里。我的研究领域之一是罕见遗传病。虽然名为 “罕见”,但实际上,罕见遗传病加起来相当普遍。全球约有 3 亿人患有罕见疾病。更重要的是,这些人往往需要经历漫长的诊断过程,可能需要数月甚至数年才能确诊。
我们正在努力开发更好的计算工具和方法来帮助我们 理解和治疗这些疾病。我们在学术环境中开展工作,深入研究罕见疾病及其病因,希望能推动这些疾病的医疗保健发展。
评估罕见疾病非常困难,因为它 需要专业的医学领域知识,还需要对生物医学数据进行系统性推理。我们认为 O1 模型强大的推理能力 正好可以解决这个问题。
John:这很有道理。我们的大型语言模型拥有领域知识,而我们的 O1 模型是强大的推理器,因此强化微调似乎是解决这个问题的理想计算方法。您能否详细介绍一下您正在使用的数据集?
罕见疾病数据集:来自数百篇科学文献
Justin:这是我们团队与德国 Charité 医院、Peter Robinson 实验室以及 Monarch Initiative 合作的成果。我们从 数百篇关于罕见疾病的案例报告的科学文献 中提取了疾病信息。我们将这些信息整理成患者存在和不存在的体征和症状列表,当然还包括他们患有的疾病以及发生突变的致病基因。
John:我明白了,所以您和医生们正在尝试根据患者的症状来确定是哪个基因发生了突变并导致了这些症状。
Justin:没错。我们一直在与 OpenAI 团队合作,训练 O1 模型更有效地推理疾病的成因。
强化微调实战:O1 Mini 性能超越 O1
John:太不可思议了,谢谢你,Justin。现在我们将向您演示强化微调的实际应用。剧透一下,我们将使用 O1 Mini,并让它 在此任务上的性能超过 O1。这意义重大,因为 O1 Mini 比 O1 更小、更快、成本更低。
我们将使用 Justin 的数据集,向您展示如何大幅提高 O1 Mini 在这项任务上的性能。这项任务的目标是:在给定症状列表的情况下,预测哪个基因可能是导致遗传疾病的原因。
为了让大家更好地理解这个过程,我们将首先介绍用于训练模型的数据集和用于评估模型的评分器。然后,我们将 在 OpenAI 的训练基础架构上启动训练任务。最后,我们将评估生成的微调模型,看看它与基础模型相比有哪些改进。
首先,我们将进入 OpenAI 开发平台并创建一个新模型。一年多来,我们一直在进行监督微调。现在,我们将选择 强化微调。我们将选择 O1 作为基础模型。接下来,我们需要上传训练数据集。
训练数据集是 JSONL 文件,其中每一行都是您希望模型学习的示例。Justin 和他的同事们收集了大约 1100 个示例 的数据集。我将上传这个数据集。
为了让大家更好地理解这个数据集,我们将放大一个单独的数据点。这就是它的样子,其中包含三个重要信息。首先是 病例报告,是对患者及其症状的描述。我们可以看到患者是一位 51 岁的女性,未指明发病时间,我们还列出了一系列症状,如眼距过宽和甲状腺功能亢进。正如 Justin 所说,我们还列出了 不存在的症状,这有助于模型排除那些它可能误认为是致病基因的基因。
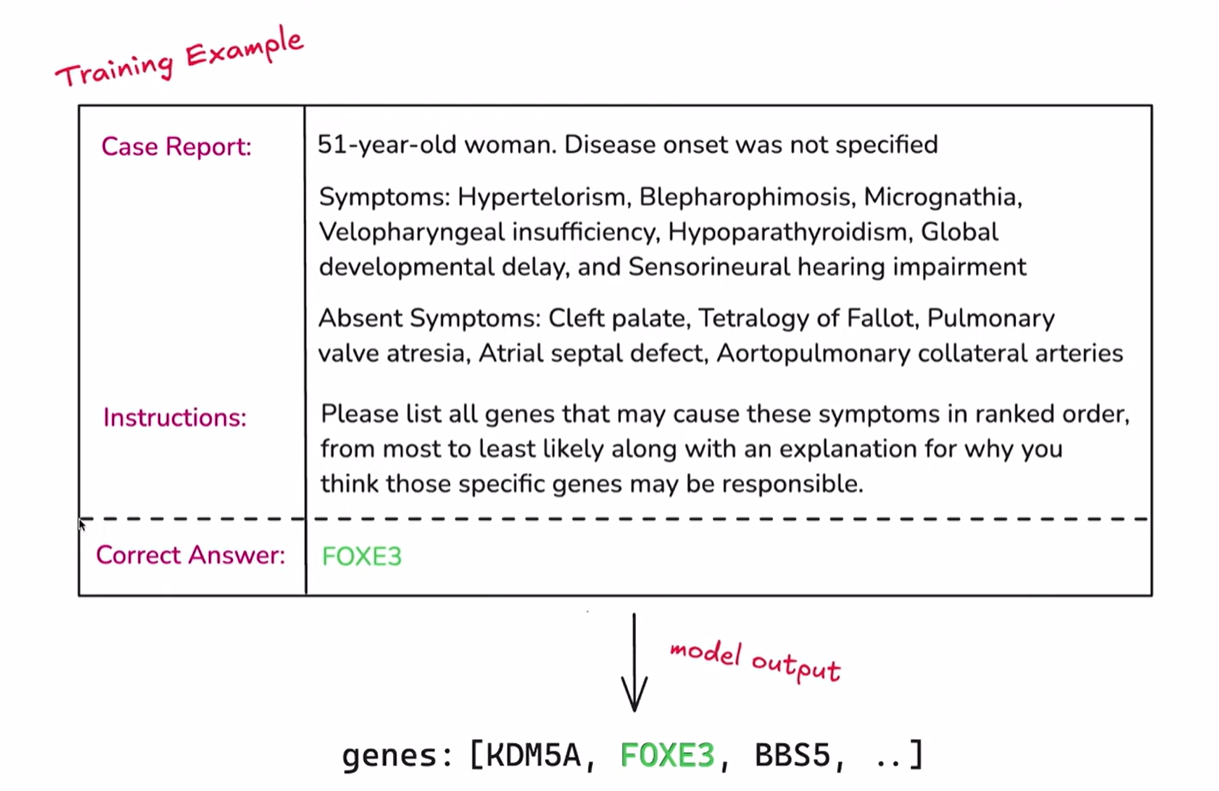
接下来是 指令。我相信观看直播的朋友们都比较熟悉提示了。我们只是在告诉模型我们希望它完成的任务。给定症状列表和病例报告,您能否列出所有您认为可能导致遗传疾病的基因? 我们还要求它提供解释。
最后是 正确答案。这是我们已知的致病基因,但在训练过程中我们不会向模型透露这个信息。否则就相当于作弊了。我们只在内部使用正确答案来评估模型的输出。
这确实是一个相当困难的任务。我肯定回答不了这个问题。
没错,我们已经取得了很大的进步,而不仅仅是让模型数 “草莓” 这个词中有多少个 R。
现在,当我们向模型提供提示、病例报告和指令后,模型会输出这样的结果,即它认为可能致病的基因列表。重要的是,这些基因是按可能性排序的,第一个基因是模型认为最有可能致病的基因,第二个基因是模型认为第二有可能的基因,依此类推。
接下来,我们需要上传 验证数据集。验证数据集的格式与训练数据集相同,但 正确的基因没有重叠。这意味着 模型无法作弊,它必须学会从训练数据集中归纳总结,并将其应用于验证数据集。
明白了。那么强化学习在哪个环节发挥作用呢?您之前提到了评分,这是其中的一部分吗?
问得好。评分是由我们引入的 “评分器” 完成的。评分器的原理很简单:它会获取模型的输出和正确答案,进行比较,并返回 0 到 1 之间的分数。0 分表示模型完全答错,1 分表示模型完全答对。您也可以给出部分分数。
针对这项任务,我们设计了这样一个评分器。它会获取正确答案以及模型的输出(即基因列表),并生成一个分数。在这个例子中,FOXP3 是正确答案。它在基因列表中排名第二,因此获得了 0.7 分。
我明白了。也就是说,如果模型将 FOXP3 排在第一位,它就能得到 1 分。
没错。模型预测的基因在列表中越靠后,得分就越低,逐渐衰减到 0。
不错,很有道理。但如果我的任务不是对排名列表进行评分呢?我们是否还有其他更通用的评分器?
当然。我们提供了一系列评分器,我们认为它们可以有效地涵盖您在进行强化微调时可能遇到的各种情况,并且我们还在不断添加新的评分器。未来,我们还将允许用户自定义评分器。
例如,上传一个 Python 文件来自定义评分规则。
太棒了。我们已经定义了训练数据集和验证数据集。现在,让我复制评分器。OpenAI 允许您通过设置超参数来自定义微调,但我们已经设置了一些非常好的默认值,所以我直接点击 “创建” 即可。
现在,训练任务已经启动。您只需要提供数据集和评分器,也就是您具备专业知识的部分。接下来,OpenAI 强大的强化学习算法和分布式模型训练系统 会接管一切,为您定制一个适用于您特定用例的前沿模型。
也就是说,作为用户,我只需要提供数据集和评分器,剩下的就交给 OpenAI 了。
没错。强化微调任务的运行时间 从几个小时到几天不等,所以我们现在来看一个我本周早些时候在相同数据集上运行的训练任务的结果。
这是我本周早些时候运行的任务。它已经成功完成,并生成了一个微调模型。我想重点关注一下 验证奖励分数。这是评分器在验证数据集上的平均得分,以及它在整个微调过程中的变化情况。我们可以看到,分数一直在上升。正如我们之前所说,由于训练数据集和验证数据集之间没有基因重叠,这意味着模型确实学会了泛化我们的任务,而不仅仅是简单地记忆症状并将其映射到基因。
虽然结果令人欣喜,图表也呈现出我们期望的上升趋势,但如果能更直观地了解模型在微调过程中是如何改进的就好了。接下来,我们就来深入分析一下。
现在,我们将进入 评估仪表板,这是我们今年早些时候在开发者平台上推出的一个产品。这里有很多数字,不用担心,我们会一一讲解。
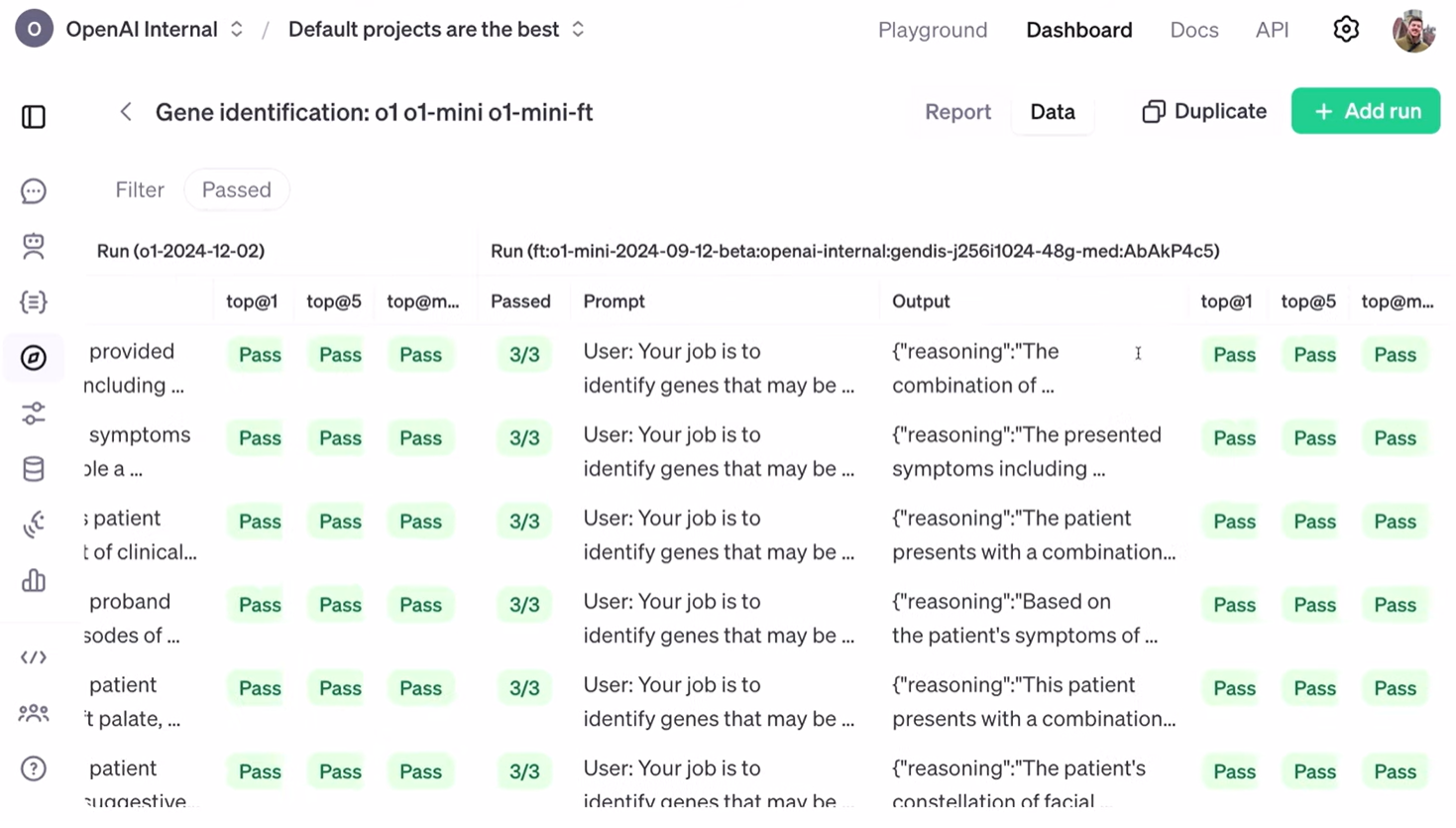
我在这里设置了三个不同的运行结果。第一个是 我们昨天发布的 O1 模型 的运行结果。第二个是 O1 Mini 的运行结果,它是我们微调的起点。最后一个是 强化微调后的 O1 Mini 的运行结果。
我们已经看到奖励分数的上升趋势,但这对这项任务来说究竟意味着什么呢?我设置了三个不同的评估指标。第一个是 “top at one”,表示正确答案在模型输出列表中排名第一的频率;第二个是 “top at five”,表示正确答案出现在模型输出列表的前五位中的频率;最后一个是 “top at Max”,表示正确答案是否出现在模型的输出列表中。
从 “top at one” 指标来看,O1 Mini 在大约 200 个样本的数据集上取得了 17.7% 的成绩。O1 的成绩更好,达到了 25%。而经过强化微调的 O1 Mini 则取得了 31% 的好成绩!
太棒了!我截取了这张图的屏幕截图,并把它上传到 ChatGPT,让它生成了一张圣诞主题的图表。这张图表清晰地展示了这九个数字。您可以看到 O1 Mini、O1 以及强化微调后的 O1 Mini 在 “top at one”、“top at five” 和 “top at Max” 三个指标上的表现。强化微调后的 O1 Mini 的性能最佳,用红色虚线表示。

Justin,您如何看待这些结果?
我认为这是一个非常 impressive 的结果,尤其是验证数据集上的提升,因为这意味着 模型正在学习一些关于如何推理此类数据的一般性知识,这非常令人兴奋。您可能会问,与现有的生物信息学工具相比,这个模型的表现如何?我目前还没有进行直接的比较,因为通常情况下您需要提供基因组测序数据,而我们这里没有包含这些数据。但模型能够基于不完整的症状列表进行开放式查询,这一点非常新颖,也很令人兴奋。
模型输出:不仅提供答案,更重要的是推理过程
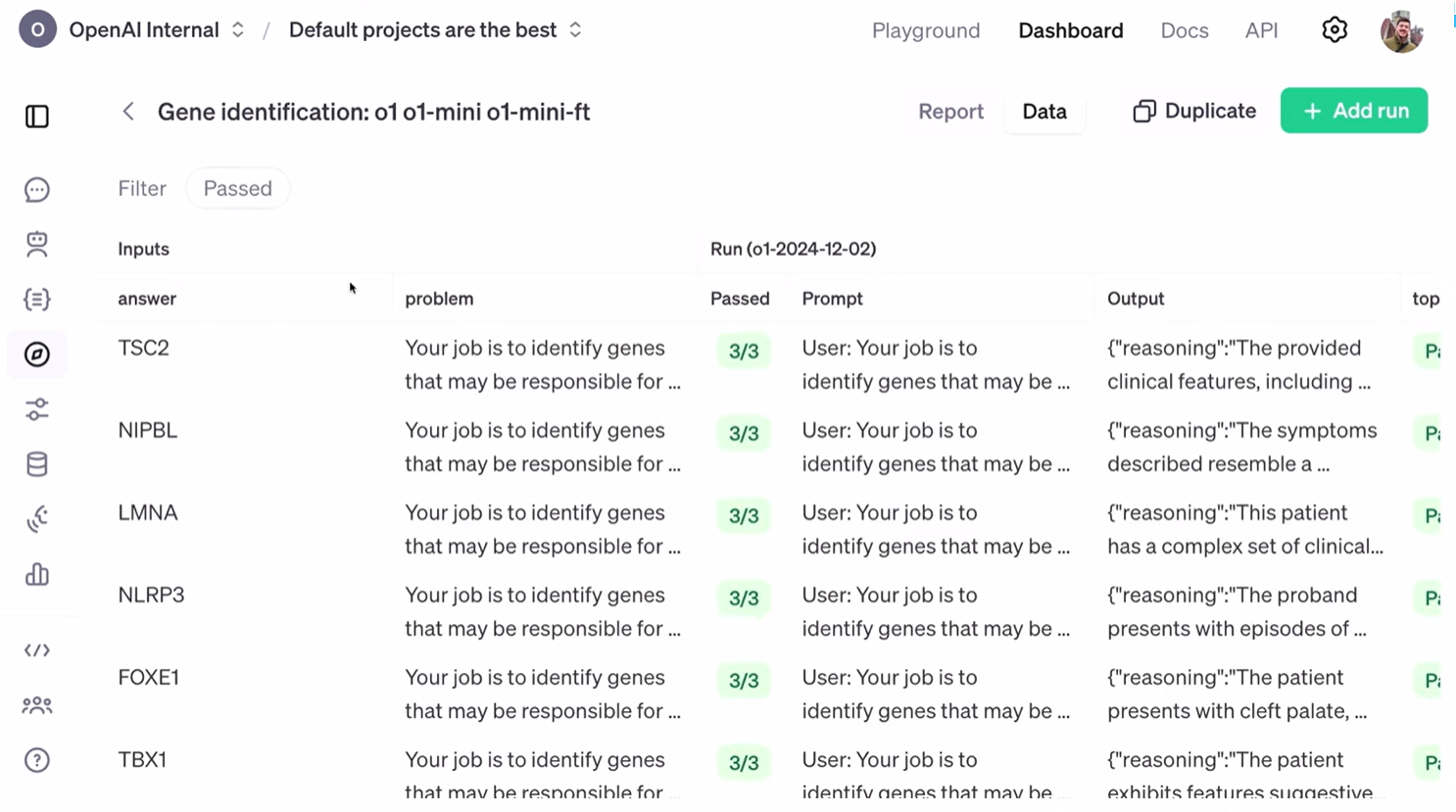
很好。以上是汇总统计数据,现在让我们来看看模型的实际输出。我将打开 “数据” 选项卡,并筛选出那些通过测试的案例。这是我们提供给模型的输入。正如 John 所说,我们的目标是 识别那些可能导致一组观察到的症状的基因。我们要求模型输出一个字典,其中包含一个字符串来解释它选择这些基因的理由,当然还有按顺序排列的基因列表。最后,我们还有症状列表。
这位患者表现出脑室下结节、癫痫发作等症状。然后,我们运行了模型。这是 O1 模型,而这是强化微调后的 O1 Mini 模型。我们将输入提供给模型后,得到了我们之前描述的字典形式的输出。
“推理:脑室下结节、癫痫发作、皮质块茎的组合表明了这种复合体,这种复合体通常是由这些基因的突变引起的。” 模型还列出了其他几个可能的基因,并最终确定 TSC2 是最有可能的候选基因。如果我们回头查看答案,就会发现 TSC2 确实是正确答案。因此,模型在 “top at one”、“top at five” 和 “top at Max” 三个指标上都通过了测试。
Justin,您认为这样的模型输出对您来说有帮助吗?
当然。模型的推理过程 对我来说特别有帮助,这是一个很大的贡献,基因的排名列表 也非常有用。即使正确答案不是排名第一,我也可以查看所有可能性。强化微调能够提升模型预测的准确性,这一点也很棒,正确答案的排名越来越靠前了。
这真是令人欣慰。Justin,从更宏观的角度来看,强化学习对您的研究领域有哪些影响? 您能谈谈一些趋势吗?
强化学习:生物信息学领域的未来趋势
我认为在研究领域,人们对 将这些模型应用于此类任务 非常感兴趣。就目前的情况来看,短期内最佳的解决方案可能是将现有的生物信息学工具与 O1 等模型结合起来。我们在描述这些模型的优势以及如何使用微调等工具来提升模型性能方面已经取得了很大的进步。
就像我说的,目前还没有合适的基准来比较这两者,但毫无疑问,我们在如何利用这些模型来理解疾病方面取得了显著的进展。从更广泛的意义上来说,如何将这些模型整合到工作流程中,最终改善医疗保健,这也是一个重要的研究方向。
强化微调:通用技术,无限可能
太棒了,谢谢你,Justin。虽然我们今天展示的是强化微调在科学研究中的应用,但它实际上是一项 通用技术。我们在生物化学、AI 安全、法律和医疗保健等领域的数据集中都看到了 promising 的结果。我们可以想到还有很多其他的应用场景,相信大家也能想到更多。这就是为什么我们今天要扩展 Alpha 计划,让更多人能够在对他们最重要的任务上 突破 O1 模型的极限。
我们一直在与一小部分值得信赖的合作伙伴合作测试强化微调,今天我们将通过 “强化微调研究计划” 来扩展 Alpha 测试的范围。该计划非常适合那些与专家团队合作处理复杂任务,并认为 AI 助理可以提供帮助的组织。如果您有兴趣申请参与这个项目,可以在直播描述中找到申请链接。正如 Mark 之前所说,我们计划在明年年初正式发布强化微调。
我们非常期待看到大家使用强化微调来做些什么。作为一名研究人员,没有什么比看到我们的模型被应用于 推动现实世界中的科学知识发展 更让我们高兴的了。
圣诞主题笑话时间
你今天有笑话要讲吗?嗯,碰巧我有。呃,由于它已成为一种传统,我有一个圣诞主题的笑话。所以,你知道,我们住在旧金山,自动驾驶汽车风靡一时,实际上,圣诞老人也一直在尝试加入其中。嗯,他正在尝试制造自动驾驶雪橇 (self-driving sleigh),但由于某种原因,他的模型总是无法识别树木,雪橇左右撞到树木。嗯,你们猜猜为什么?
不,“他没对他的雪橇进行‘微调’,而是进行了‘树木微调’……”("He didn't pine-tune his...")
谐音梗:
-
"pine-tune" 的发音和 "fine-tune" (微调) 非常相似。
-
"pine" 是松树的意思,圣诞树通常就是用松树做的。
好的,好的。嗯,请下周加入我们;我们还有更多东西要分享。谢谢!
SEO 技术优化和 AI 强化微调
在筋斗云 SEO 中,已经落实的 AI SEO 微调模型已经有这些,为我们的 SEO 优化工作提升质量:
-
定制 SEO 内容生成模型:
-
训练数据: 收集了许多高质量的 SEO 内容样本,包括文章、标题、描述等,并将其整理成符合强化微调要求的格式。
-
评分器: 根据 SEO 最佳实践,设计一个评分器来评估模型生成内容的质量,例如关键词密度、内容相关性、可读性等。
-
微调模型: 使用收集到的数据和设计的评分器,对 O1 模型进行强化微调,使其能够生成更符合 SEO 要求的内容。
-
-
自动化 SEO 任务:
-
自动生成 SEO 报告: 自动生成各种 SEO 报告,例如网站流量分析报告、关键词排名报告、竞争对手分析报告等。
-
自动优化网站内容: 自动优化网站内容的标题、描述、关键词密度等,使其更符合搜索引擎的排名规则。
-
自动检测和修复 SEO 问题: 例如 broken links、duplicate content 等,并提供修复建议。
-
-
开发 SEO 工具:
-
内容优化工具: 根据目标关键词和用户搜索意图,自动生成高质量的 SEO 内容,并提供优化建议。
-
网站分析工具:更深入地分析网站流量数据、用户行为数据等,并提供更具洞察力的 SEO 建议。
-

Reinforcement Fine-Tuning—12 Days of OpenAI: Day 2 英文字幕整理版本
Hi everyone, my name is Mark, and I lead research at OpenAI. Yesterday, we took O1 out of preview and launched it in ChatGPT. We're soon going to launch it in the API as well.
If you haven't been following O1, it's our latest series of model improvements that allow the models to think for a while before responding. Today, we're excited to preview our latest advancement in model customization: reinforcement fine-tuning. This will let users fine-tune O1 on their own datasets.
Again, this isn't standard fine-tuning; this is reinforcement fine-tuning, which leverages the reinforcement learning algorithms that took us from advanced high school level to expert PhD level. This is a preview of something we'll launch publicly next year, but if you're a university, researcher, or enterprise, we'll share information on how you can access our program later.
Why would you want this? Well, it allows you to take your golden datasets and turn them into unique offerings with the same magic we have, but for your own users and customers. I'll let John, Julie, and Justin say a little bit more.
John: Hello everyone, my name is John Allard, and I'm an engineer here at OpenAI.
Julie: Hi everyone, I'm Julie W, a researcher here at OpenAI.
Justin: I'm Justin Ree, a computational biologist at Berkeley Lab.
John: Today, we're excited to introduce a new way of model customization for our O1 series: reinforcement fine-tuning, or RFT for short. For the first time, developers, researchers, and machine learning engineers will be able to use reinforcement learning to create expert models capable of excelling at specific tasks within their domain.
We believe any field requiring deep expertise in AI models stands to benefit. So, if you work in law, finance, engineering, insurance, this is for you. For example, we recently partnered with Thomson Reuters to use RFT to fine-tune O1 Mini to be a legal assistant in their co-counsel AI. This tool assists their legal professionals in accomplishing some of their most analytical workflows.
Julie: Some of you will be familiar with the supervised fine-tuning API we launched early last year. Supervised fine-tuning is powerful when you're trying to get the model to replicate features it finds in input text or images. This is great if you want to change the tone, style, or response format of the model.
Now, with reinforcement fine-tuning, or "reindeer-forcement" fine-tuning, as I should say, it's different. You're not just teaching the model to mimic its inputs; you're teaching it to reason in entirely new ways over custom domains.
When the model sees a problem, we give it space to think through it, and then we grade the final answer. Using reinforcement learning, we reinforce lines of thinking that led to correct answers and disincentivize those that led to incorrect answers. With as little as a few dozen examples, the model will learn to reason in new and effective ways.
John: That's crazy that you can do that with just 12 examples. That's not something you can do with regular fine-tuning.
Julie: Exactly! In the world of large language models, a few dozen examples is basically nothing.
John: For the first time, our model customization platform will support reinforcement learning. Notably, this is the same technique we use internally at OpenAI to train our frontier models like GPT-4 and the O1 series. One area with many exciting applications is scientific research. But don't just take our word for it; that's why we're joined today by Justin Ree. Justin is a researcher at Berkeley Lab, and one of his areas of study is using computational methods to understand the genetic causes of rare diseases.
Justin, thank you so much for being here. Do you mind telling us a little bit more about your research and how reinforcement fine-tuning might help?
Justin: Thanks, it's great to be here. One of my research areas is rare genetic diseases. Contrary to the name, rare genetic diseases are actually quite common collectively. We're talking about 300 million people globally who suffer from a rare disease. What's more, these people often have a long diagnostic odyssey of months to years before they find out about their condition.
We're working on better computational tools and methods to help us understand and treat these diseases. We do our work in an academic setting, learning more about rare diseases and their causes, with the hope of advancing healthcare for these folks.
Assessing rare diseases is hard because you need expert domain knowledge about the medical side of things, and you also need systematic reasoning over biomedical data. This is where we think the O1 model can really help us out with its reasoning capabilities.
John: That makes a lot of sense. Our large language models have domain knowledge, and our O1 models are strong reasoners, so it seems like a good computational method for addressing this. Can you tell us a little bit more about the datasets you're using?
Justin: This was a collaborative effort between our group, Charité Hospital in Germany, Peter Robinson's lab, and the Monarch Initiative. We extracted disease information from hundreds of scientific publications that were case reports about rare diseases. We curated the information into lists of signs and symptoms that were present and absent in the patient, and then, of course, the disease they had and the causative gene that was mutated.
John: I see, so you and some doctors are trying to figure out, given a patient's symptoms, what gene might have mutated to cause those symptoms.
Justin: That's right. We've been working with the OpenAI team to train the O1 models to reason more effectively about the causes of disease.
John: Incredible, thank you, Justin. We're now going to give you a preview of reinforcement fine-tuning at work. Not to steal any thunder, but we're going to take O1 Mini and make it exceed the performance of O1 on this task. This matters because O1 Mini is a smaller, faster, and cheaper model than O1.
Using Justin's dataset, we're going to show you how to drastically improve the performance of O1 Mini on this task, where, given a list of symptoms, you're trying to predict which gene might be responsible for the genetic disease.
To give an overview of this process, we're going to start by looking at the datasets used to train the model and the graders used to evaluate it. Then, we're going to launch a training job on OpenAI's training infrastructure. Finally, we'll evaluate the resulting fine-tuned model to see how it's improved over the base model.
To start, we're going to jump over to the OpenAI development platform and create a new model. We've had supervised fine-tuning for a bit over a year now. We're going to select reinforcement fine-tuning. We'll select O1 as the base model. Now, we need to upload a training dataset.
Training datasets are just JSONL files, where each line is an example you want the model to be trained on. For this case, Justin and his colleagues assembled a dataset of about 1,100 examples. I'll go ahead and upload that.
To get a feel for how this dataset works, we'll zoom in on an individual data point. This is what it looks like, and there are three important things here. First is the case report, a description of the patient and their symptoms. We see the patient was a 51-year-old woman, the disease onset was not specified, and we have a list of symptoms like hypertelorism and hyperthyroidism. As Justin said, we have the absent symptoms, which helps the model rule out genes it might otherwise think are responsible.
Next, we have the instructions. I'm sure if you're watching this live stream, you're familiar with prompting. We're just prompting the model for what we want it to do for this task. Given the list of symptoms and the case report, can you list all the genes you think might be responsible for the genetic disease? We also ask it to provide an explanation.
Finally, we have the correct answer. This is the gene that we happen to know is responsible, but we're not showing this to the model during training. That would be cheating. We're using it internally to grade the model's outputs.
This is a pretty hard task. I definitely have no hope of answering this question.
Yeah, we've come a long way from just trying to count the number of Rs in the word"strawberry."
Now, when we give the model this prompt, this case report, and these instructions, the model will output something like this, which is a list of genes it thinks might be responsible. Importantly, the genes are in sorted order, where the first gene is the one it thinks is most likely to be responsible, the second one is the second most likely, and so on.
We'll hop back over. Next, we need to upload some validation data. Validation data is in the same format as the training data, but there's no overlap in the correct genes. This means the model can't cheat; it has to generalize from the training dataset to the validation dataset.
Gotcha. So, where does the reinforcement part come in? You know, we talked about grading. Is that part of the process here?
That's a good question. Grading is done by these"graders"that we're introducing. Graders are simple: they take the output from the model and the correct answer, compare them, and return a score between zero and one. Zero means the model didn't get the answer correct at all, and one means it got it completely correct. You can also give partial credit.
For this task, we have a grader that looks like this. It takes the correct answer and the output from the model, which is the list of genes, and produces a score. In this case, FOXP3 is the correct answer. It was second in the list of genes, so it gets a score of 0.7.
I see. So, if it had instead said FOXP3 was first in the list, it would have gotten a grade of one.
Exactly. And as it gets further down the list, the score gradually decays to zero.
Nice, makes sense. But what if I have a task that isn't grading a ranked list? Do we have other graders that are more general?
Yeah, we're supplying a collection of graders that we think effectively cover the space of possible intents you might have while doing reinforcement fine-tuning, and we're always adding more. Eventually, we're going to let you define your own graders.
Maybe like upload a Python file or something and do some custom grading.
Cool. So, we've defined our training dataset, we've defined our validation dataset. Let me go ahead and copy in the grader. OpenAI allows you to customize these fine-tuning runs by setting hyperparameters, but we've set some pretty good defaults, so I'm just going to click "create" here.
Now, what this is doing is kicking off a training job. The cool thing is that you bring the dataset and the grader, which are the places where you have domain expertise. Then, you leverage the full power of OpenAI's reinforcement learning algorithms and our distributed model training stack to customize a frontier model for your use case.
So, as a user, I just bring my dataset and grader, and OpenAI takes care of everything else.
Exactly. Reinforcement fine-tuning jobs can take anywhere from a few hours to a few days to run, so we're going to jump over to a job I ran earlier this week on the same dataset, just so we can see the results.
I have this job that I ran earlier this week. It completed successfully and produced a fine-tuned model for us. There's one thing I want to look at, which is the validation reward score. This is the average score from the grader on the validation dataset and how it changed over the course of the fine-tuning run. We can see that the score is going up. As we said earlier, since there's no overlap in genes between the training dataset and the validation dataset, it means the model really learned to generalize on our task. It wasn't simply memorizing symptoms and mapping those to genes.
While this is cool, and the chart goes up and to the right, which is what we like to see, it'd be nice if we could get a better feel for how the model has actually changed during the fine-tuning process. We'll take a closer look at that now.
All right, so we're going to pop over to the evaluations dashboard, a product in our developer platform that we launched earlier this year. There are a lot of numbers, but don't worry, we're going to go through all of them.
I've set up three different runs here. The first one was a run against our O1 model, which we released yesterday. The second was against O1 Mini, the starting point of our fine-tuning job. And finally, the reinforcement fine-tuned O1 Mini.
We looked at the reward going up and to the right, but what does that actually mean for this task? I've set up three different evaluations to assess that. The first one is"top at one,"which is how often the correct answer is the very first item in the list."Top at five"is how often the correct answer is in the top five elements of the list. And finally,"top at max," did we at all put the right answer in our list?
Looking at "top at one," we can see that our starting point, O1 Mini, got 17.7% on our dataset of about 200. O1 got 25%, so it's doing better. But then our fine-tuned O1 Mini got 31%!
Awesome! I took a screenshot of this and put it into ChatGPT and asked it to make me a Christmas-themed plot. Here's a nice visualization of those nine numbers. You can see our starting point, O1 Mini, across"top at one,""top at five," and "top at max," our O1 model, and then finally our best performing model, which is this O1 Mini fine-tuned here in the dotted red line.
So, looking at these results, what do you think, Justin?
Well, I think this is pretty impressive performance, especially the increase in the validation data because that implies the model is learning something generally about how to reason over this kind of data, which is exciting. An obvious question you might ask is how this is doing compared to existing bioinformatics tools. I don't really have an apples-to-apples comparison because typically you would provide genomic sequencing data, and we haven't included that here. But the open-ended querying of models here over incomplete symptom lists is new and exciting.
Great. So, these are aggregate statistics, but let's look at the actual model responses. I'm going to pop over to this "data" tab. Let's filter by the passes. Here's the input that we're giving to the model. The problem, as John described, is to identify genes that may be responsible for a set of observed symptoms. We asked the model to output a dictionary containing a string that explains why it picked these genes and, of course, the genes themselves in ranked order. Finally, we have the symptom list as well.
This patient presented with subependymal nodules, seizures, and a couple of other things. We then run our models. This was our O1 model, and this one, our fine-tuned O1 Mini model. We gave it that input, and now the output is this dictionary that we described earlier.
"Reasoning: The combination of subependymal nodules, seizures, cortical tubers are indicative of this complex, which is commonly caused by mutations in these genes." It lists a couple of other potential ones and then says TSC2 is the most likely candidate. And if we scroll back over to our answer, we'll see that TSC2 is in fact the correct answer. So that allowed us to get a pass on"top at one,""top at five," and "top at max."
So, looking at this output, Justin, is this a useful output for the model to be giving back?
Absolutely. It's particularly useful to see the model's reasoning, and that's a big contribution here, and also the ranked list of answers. Even if the correct answer isn't first, you can look at all the possibilities. It's also great to see the fine-tuning improves the performance in the rank list of possible answers, so that right answer is getting closer to one.
That's gratifying. Justin, zooming out a little bit, how does reinforcement learning shape your field? Can you talk about some trends?
I think there's a lot of interest in the research community in using these models for these kinds of tasks. The feeling for this use case is that the best solution in the near term is probably a hybrid solution between existing bioinformatic tools and these models like O1. This is excellent progress in characterizing the strengths of these models and how we can use tools like fine-tuning to improve performance.
Like I said, there's not really a comparable benchmark to compare the two, but it's definitely progress in how we can use these models to understand disease, and in a larger sense, how we can incorporate these models into a workflow that will eventually improve healthcare.
Amazing, thank you, Justin. So, while we've just shown you an exciting application of reinforcement fine-tuning in scientific research, this is a general-purpose technique. We've seen promising results in datasets from biochem, from AI safety, from legal, and from healthcare as well. We can think of hundreds of more examples or tasks that we can use this model on, but we know that you can probably think of many more. That's why we're so excited to be expanding our Alpha program today to enable more people to push the boundaries of our O1 models on the tasks that matter most to them.
We've been working with a small group of trusted partners to test out reinforcement fine-tuning, and today we're expanding Alpha access via what we're calling the"Reinforcement Fine-Tuning Research Program."This program is ideal for organizations working on complex tasks with teams of experts who think they might benefit from AI assistance. If you're interested in applying for one of these limited spots, you can find a link to the application in the description of this live stream. As Mark said, we plan on launching this product, reinforcement fine-tuning, publicly early next year.
We're truly excited to see what you do with reinforcement fine-tuning. Speaking as a researcher, there's nothing that makes us happier than seeing our models being adapted and used to advance science and knowledge in the real world.
Do you have a joke for us today?
Well as so happens I do. Uh, as it's become a tradition, I have a Christmas-themed joke. So, you know, we live in San Francisco, self-driving vehicles are all the rage, and actually, Santa's been trying to get in on this too. Um, he's trying to make a self-driving sleigh, but for some reason, his models just keep not identifying trees, and the sleigh is hitting trees left and right. Uh, do you guys have any guesses why?
[pause]
No, he didn't... he didn't pine-tune his...
[laughter]
Okay, all right. Um, please join us next week; we'll have a lot more to share. Thank you!

